Juan Poyatos, Elena Camacho, Luciano Marcon and Fernando Casares
From June 2nd to June 7th, 2024, the ALiS team had the privilege of participating in a stimulating workshop hosted at the Institute of Mathematics of the University of Granada (IMAG) in Granada, Spain. Organized as a Banff International Research Station (BIRS) workshop, this event brought together leading researchers and experts from around the world to explore the dynamics of biological patterning and development with mathematical modeling. All the members of the ALiS team were at the event and gave a talk. The workshop was attended only by the speakers, making it a cozy meeting among interdisciplinary mathematical enthusiasts interested in mathematics, biology, and computational modeling.
The workshop titled “From Evolution to Bioengineering of Biological Patterning Mechanisms – Mathematical Advances and Challenges” aimed to explore the fundamental principles governing biological patterning and development across various organisms, from single-celled cyanobacteria to complex multicellular organisms like plants and animals. BIRS workshops are known for their interdisciplinary approach, bringing together experts from diverse fields to tackle complex scientific challenges.
Program Overview:
Day 1 (June 3): The workshop kicked off with a series of insightful talks about the main aspects of biological patterning. Eric Siggia opened the day with his presentation on “Geometry and Genetics,” proposing geometric descriptions as a suitable formalism to encode dynamic genetic regulation in biological systems. James Sharpe followed with “A periodic table of patterning mechanisms?” where he discussed systematic theoretical framework for exploring and understanding the diverse range of patterning mechanisms based on the topology of gene regulatory networks. Jeremy Green discussed “Morphogen cocktails and mechanical symmetry-breaking,” shedding light on the synergistic interplay between morphogens and mechanical forces in shaping tissue patterns showing preliminary work on convergent extension movement in Xenopus explants. Kate McDole’s talk, “How the embryo gets its shape,” offered illuminating insights into the mechanisms underlying mouse gastrulation and foregut development through adaptive light-sheet live-imaging techniques. She concluded the talk proposing that all the data produced by her lab will be integrated in a “virtual computational embryo”. Finally, Michel Milinkovitch presented “How reptiles got their looks,” exploring the patterning of skin colors and appendages in vertebrates with self-organizing models. This was an amazing talk, with stunning visuals and modeling of pigment patterning in lizards and skin folding in crocodiles. The afternoon session featured a series of compelling talks on vascular biology and tissue remodeling. Kristina Haase’s presentation, “Vasculature as a dynamic and complex network by in vitro design,” emphasized the significance of microfabrication and tissue engineering in studying vascular behavior in controlled environments. Sara Nunes Vasconcelos discussed “A multipronged strategy for uncovering the blueprint to successful re-vascularization,” shedding light on the challenges of generating functional vasculature for organ regeneration and proposing innovative solutions. Noelia Grande Gutierrez introduced “Computational multiscale modeling to determine mechanical and biochemical environments for tissue remodeling,” showcasing the potential of computational modeling in understanding tissue dynamics and its practical implications in disease contexts. Maria Holland explored “Patterns of cortical thickness and 3D curvature in human development and mammalian evolution,” studying the mechanical mechanisms underlying the intricate folding patterns of the brain cortex across different developmental stages and species.
Day 2 (June 4): Day two featured a comprehensive exploration of mathematical modeling and analysis in the context of biological patterning and morphogenesis. Fernando Casares commenced the day with “The problem of organ SIZE,” addressing the fundamental question of how the Drosophila eye achieves its characteristic size during development via a feedback control on progenitor apoptosis. Anqi Huang’s presentation, “GRN evolution, developmental robustness and morphological innovation,” focused into the evolutionary dynamics of gene regulatory networks and their implications for developmental robustness. Roman Vetter from Iber’s lab discussed “Modeling the precision of tissue patterning with noisy morphogen gradients” discussing insights into the different mechanisms that could help to improve the precision of morphogen gradients. Alejandro Torres-Sánchez presented “Computational modeling of fluid interfaces,” highlighting the utility of computational approaches in understanding the dynamics of fluid systems relevant to biological patterning. Timothy Saunders elucidated “Quantifying 4D cell morphodynamics during skeletal muscle formation,” providing a detailed analysis of cell shape changes during tissue morphogenesis in Zebrafish. The afternoon session begun with Luis María Escudero talk “How scutoids explain 3D epithelial organization,” shedding light on the geometric principles underlying epithelial cell packing in three-dimensional space. Herve Turlier presented innovative methods to extract physical information from fluorescence microscopy images with computational models of tissue mechanics and AI. Tatyana Gavrilchenko explored the structure-function relationship in the insect respiratory system, offering insights into network growth and oxygen distribution. Alexandria Volkening gave an interesting talk titled “Modeling and quantifying cell behavior in biological patterns” tackling the problem of Turing pattern quantification using topological data analysis techniques. Finally Adrian Buganza-Tepole investigated the mechanobiological control of wound healing, revealing the complex interplay between mechanics and biology.
Day 3 (June 5): Jose A. Carrillo commenced the day with “Cell-cell Adhesion micro-and macroscopic models via Aggregation-Diffusion systems” presenting ways to model how cell-cell adhesion can shape tissue architecture. Vaclav Klika’s presentation on “Characteristic wave speed in reaction-diffusion systems” provided a detailed analysis of Turing pattern formation, emphasizing the importance of a characteristic patterning speed that propagate as a wave. Marta Ibanes discussed “Patterning by a diffusion-driven switch,” exploring the mechanisms underlying patterning processes driven by a diffusible signaling molecule coupled with bistability and auto-activation. Isaac Salazar-Ciudad offered thought-provoking insights into “What morphogenesis and pattern formation tells about the tempo and mode of evolution,” highlighting the interplay between developmental processes and evolutionary dynamics. Luciano Marcon concluded the morning session with “A topological atlas of multi-functional self-organizing Turing networks,” providing a comprehensive analysis of the topological properties of self-organizing networks underlying the formation of periodic patterns and traveling waves.
The day was conclude with a social dinner at the garden of the university residence Carmen de la Victoria from the University of Granada with a beautiful view on Alhambra.

Day 4 (June 6): The fourth day continued the exploration of biological patterning mechanisms across different organisms and developmental contexts. Maria Jose Jimenez delved into “On topological analysis of biological data,” demonstrating the utility of topological data analysis in uncovering hidden patterns in complex biological datasets. Jose A Langa’s presentation on “Complex networks and dynamics” stressed with “love” the need to consider systems where the position of attractors move over time as the most biologically realist scenario. Juan Poyatos discussed “The Developmental Foundations of Complex Phenotypes in C. elegans,” shedding light on the intricate interplay between developmental processes and the emergence of complex phenotypic traits, showing how data can confirm and contradict the intuitive law that “similar genotype should lead to similar phenotype”. Elena Camacho Aguilar’s presentation on “Modelling Cell State Transitions in Embryonic Development” offered insights into the dynamics of cell state transitions during embryonic development using catastrophe theory and non-autonomous transitions. This gave a more detailed explanation and introduction on the topic given by Siggia on the first day. Alexis Villars explored “Epithelial cell extrusion,” elucidating the regulatory mechanisms governing cell extrusion processes in epithelial tissues. Saúl Ares provided a more detailed discussion of the networks that controlled Drosophila eye development presented by Casares on the previous day. Finally Ben Shirt-Ediss discussed the emergence of minimal agency in protocells through computational modeling, revealing how regulatory mechanisms evolved to enable survival in changing environments.
At night the ALiS team was unleashed in Granada, to sell our scientific ideas and our carpets!

Day 5 (June 7): The final day of the workshop featured online talks by Kirsten ten Tusscher and Michael Stumpf, exploring lateral root development and equivalences between Turing pattern and positional information mechanisms, respectively.
Discussion:


The workshop concluded with a discussion session, providing participants with an opportunity to reflect on the themes explored throughout the week and engage in interdisciplinary dialogue to further advance our understanding of biological patterning and development. A central theme that emerged from the workshop discussions revolved around the concept of landscapes of potential dictating the genetic dynamics of cells during development. This metaphorical landscape represents a multidimensional space where each point corresponds to a potential cell state or fate.
James Sharpe initiated and moderated the discussion, drawing attention to an old John Reinitz’s paper where the dynamics of the GAP gene patterning system are governed by transient dynamics at the boundary between GAP gene expression, potentially stimulated by a global morphogen gradient making the decision non-autonomous .
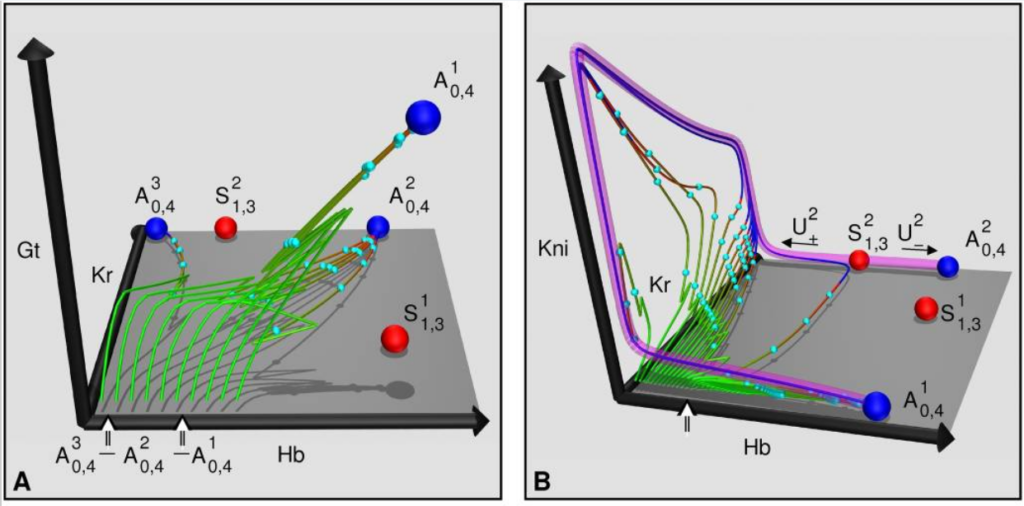
This lead speakers to discuss the contrast between autonomous and non-autonomous landscapes. In an autonomous landscape, the regulatory network within the biological system determines its own fate and therefore the trajectory of cells in the phase space. Conversely, in a non-autonomous landscape, external inputs or signals can dynamically change the landscape, affecting the trajectory of cellular development. These external influences modulate attractors pushing the system towards specific cell states or phenotypic outcomes. It was also discussed that the shape of the potential landscape is closely tied to the “geometry” of the attractors within the system. Stable attractors represent stable cell states, while unstable attractors indicate transitional states where cells are on the verge of determining their fate. The interaction between stable and unstable attractors determines the trajectories observed during development, which as proposed by Langa in many cases may be the only thing that really matters in biological complex systems, since it is very likely that attractors are moving in time and are never reached. Marcon discussed that we should define non-autonomous systems as those where time-dependent boundary conditions (or external inputs) determine part of the system dynamics. The debate also recognized a distinction between transient dynamics and non-autonomous dynamics, highlighting the complexity of development.
Additionally, speakers highlighted the importance of mathematical modeling in understanding the dynamics of potential landscapes. By creating computational models based on gene regulatory networks and signaling pathways, researchers can simulate how genes regulatory networks shape the landscape. Alternatively geometric landscape description can model the same phenomena with less parameters. These type of approaches can offer valuable insights into the factors influencing pattern formation, cell fate decisions, and the effect of developmental pathways.
During the discussion, it became apparent that a key point of debate was the extent to which biological systems should be considered autonomous or non-autonomous. For instance, Eric Siggia highlighted observations that wherever a Drosophila embryo is placed, it develops normally without external input, suggesting that in contrast to mammalian development it is an autonomous process. Sharpe emphasized that while a system like the Drosophila embryo may be considered autonomous as a whole, it can be seen as non-autonomous when dissected into parts, with each part influencing the others’ dynamics.
Overall, it was an exceptional meeting! The workshop’s format, with its relatively long duration and intimate setting, played a pivotal role in stimulating real scientific discussions among participants. We returned enriched and deeply grateful to the organizers for giving us this incredible experience.